oxen diff command with the path to your datasets.
Diff Types
Oxen.ai currently supports a TextDiff and a TabularDiff data type. TheTabularDiff data type is used to represent the differences in tabular data, such as CSV, TSV, or Parquet files. The TextDiff data type is used to represent the differences in text files, such as markdown, code, or configuration files. In the future, we plan to add support for other data types such as images, audio, and video.
Diff Command Syntax
Theoxen diff command supports multiple syntax patterns similar to Git:
oxen diff- Compare working tree with HEADoxen diff <path>- Compare specific file in working tree with HEADoxen diff <commit>- Compare commit with HEADoxen diff <commit>..<commit>- Compare two commits using range syntaxoxen diff <commit> <commit>- Compare two commitsoxen diff <commit> [--] [<path>...]- Compare commit with HEAD for specific pathsoxen diff <commit>..<commit> [--] [<path>...]- Compare two commits for specific pathsoxen diff file1 file2- Compare two local files (not in repository)
.. separator can be used to specify commit ranges, making it easy to compare different versions of your data.
Pick Your Tooling
All the functionality below is available through the 🖥️ Command Line, 🦀 Rust Library, 🐍 Python Library, as well as the 🌎 Web Interface. This guide will focus on the command line tooling, but the same principles apply to the other interfaces. Using the Oxen.ai Hub you can quickly visualize and navigate the changes in your datasets with an easy to use interface. Sign up for free 👉 here.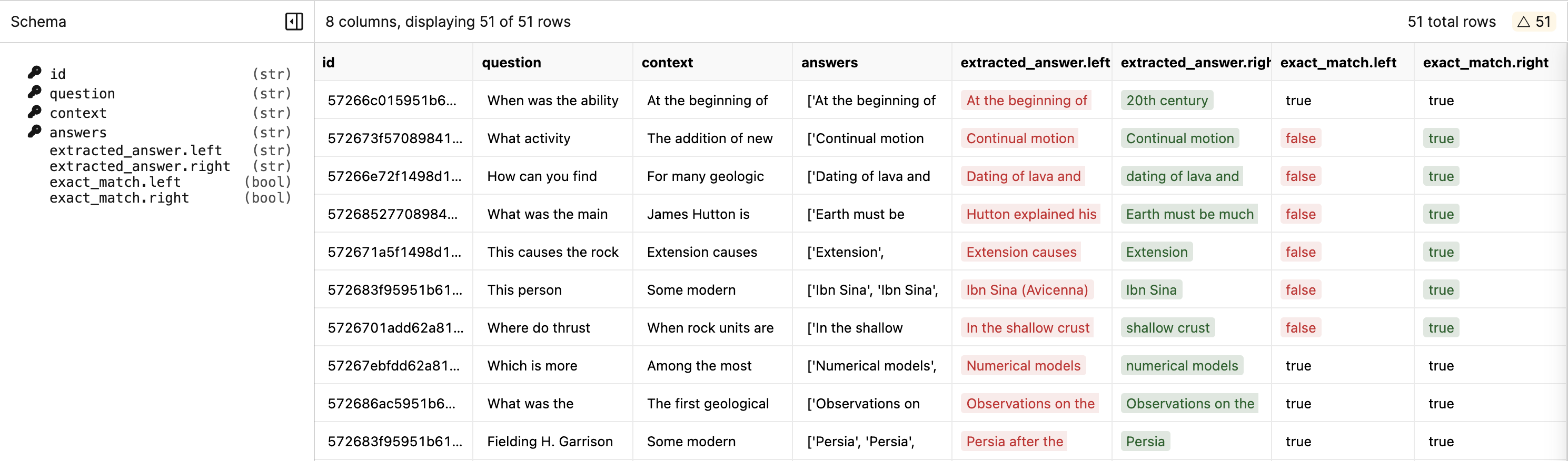
Let’s Build a Dataset
In order to demonstrate how to use theoxen diff command, we will need a dataset to work with. Imagine we are collecting a dataset for fine-tuning a Large Language Model (LLM). This dataset will have a set of prompts and a category that they belong to.
Create a new file called dataset.csv and add the following data to it.
oxen df command it is a handy tool to manipulate and inspect tabular data. You can use it with any CSV, TSV, Parquet, or line delimited JSON file.
Adding Rows
Let’s start with a completely additive workflow as if we are collecting a large datasets of prompts. Add a row to the dataset by simply appending to the file.oxen diff command. By default, Oxen compares the working tree with the HEAD commit (the last committed version).
.oxen.diff.status column to show the status of the row.
There are three possible values for the .oxen.diff.status column:
addedremovedmodified
Removing Rows
Next remove the first entry of the file to see how Oxen handles deletions. We will use thesed command with the in place flag -i to remove the first row from the file.
-i '' flag is for MacOS, if you are using Linux you can simply use -i.) Since the file is a CSV with a header row, you will need to remove the second row hence 2d.
Verify that the first row was removed by using the oxen diff command.
Modifing Rows
This is great for adding and removing rows, but what about modifying rows? Say we change thecategory of “geography” to be a more generic “trivia” category and add a new prompt to it “What is the fastest land animal?”.
Edit the datasets.csv file to look like this:
oxen diff command again, we will see the changes.
Specifying Keys
The reason that the above example treats the modified row as a new row and a removed row is because both theprompt and category columns being considered keys under the hood. oxen diff hashes the combination of keys in order to find differences in the data. The default keys are all the common columns between the two versions of the datasets.
If you have a unique identifier for each row, you can use the --keys (or -k) flag to specify the column or columns that should be used as the primary keys.
category.left and category.right, to show the old and new values.
Assumming these changes look good, you can add and commit the changes to your local repository.
Adding Columns
Adding and removing rows is great, but what about changes to the schema itself? Instead of using the prompt as a key, let’s add anid column to the dataset and use that as the key. Let’s also add an answer column to the dataset, so that we can evaluate the responses.
Update your raw csv with the new columns like so:
oxen diff command, you will see that it automatically detects the added columns and displays the new values in id.right and answer.right.
.left to show the values in columns that are now missing. If you are happy with the changes, you can add and commit the changes to your local repository.
Specifying Compares
Not only can you specify keys to narrow down the scope of what fields oxen hashes, but you can also specify columns to compare with the--compares (-c) flag. This specifies the fields oxen compares.
You can think of the keys as the fields that are hashed to create a unique id to tell if a row was added or removed. The compares are the fields that are compared to check if a row was modified. By default if you specify a single key, the rest of the columns become the compares. If you specify multiple keys, the compares are all the columns that are not keys.
To see this in action, let’s add one row, remove one row, and modify 3 existing ones to demonstrate how this works. In this case we will only modify values of the answer column.
Overwrite the dataset.csv file with the following data.
-c flag to specify that we are only interested in changes in the answer column.
-c flag comes in handy.
To see how this works, try using the -k flag on the same dataset without any compares.
(5 rows x 7 columns) which isn’t too bad, but if you have a dataset with many columns, it can quickly become overwhelming with irrelevant information. If you know where to look, you can use the -c flag to narrow down the scope of the diff.
Saving Results
The--output (-o) flag can be used to save the results of the diff to a new file. This is useful if you want to save the results of the diff to a new file for further inspection or to share with others.
diff.csv. You can then load it into a jupyter notebook, pandas, or even back into Oxen to do more analysis on the results.
Real World Example
To drive all these features home, imagine you have taken the dataset above and run it through an LLM with a prompt to get the responses. You have saved the results in a new file calledmodel_results.csv.
Below is an example script that runs the prompts through gpt-3.5-turbo and saves the results to a new file. This script uses the openai python package to interact with the OpenAI API.
process_csv_with_openai.py
dataset.csv file to get the model_results.csv file.
model_results.csv file with the oxen df command to make sure the csv was created correctly.
id, prompt, answer, and category columns as the original dataset, but it also has some additional columns such as response, is_correct, model, and inference_time.
Add and commit the model results to your local repository.
oxen diff command to see the differences.
Overwrite the model_results.csv file with the new results.
model and inference_time columns could be different for each row.
response and is_correct columns, and ignore the model and inference_time columns.
In combination with the --keys flag, you can use the --compares (or -c) flag to specify the columns you are interested in.
response and is_correct columns. We can see that the new model has a different response for the prompts 1 and 3. Diff allows us to quickly narrow down the responses that model 1 and model 2 disagree on, and which ones are correct.